publications
publications by categories in reversed chronological order. generated by jekyll-scholar.
2025
- Nature Sci. Rep.
 Normal twin PET: personalized generative modeling for confounder correction and anomaly detection in whole-body PET/CTChristian Hinge, Anders Bertil Rodell, Sven Zuehlsdorff, and 5 more authorsScientific Reports, Nov 2025
Normal twin PET: personalized generative modeling for confounder correction and anomaly detection in whole-body PET/CTChristian Hinge, Anders Bertil Rodell, Sven Zuehlsdorff, and 5 more authorsScientific Reports, Nov 2025Variable physiological [18F]FDG uptake patterns and a lack of labelled data make it challenging to automatically distinguish normal from pathological suspicious uptake in whole-body PET/CT imaging. We propose a deep learning method that generates patient-specific normal twin PET images to serve as personalized references for quantitative analysis and unsupervised detection of pathological anomalies. We developed an image-to-image generative model that synthesizes normal reference twin PET (ntPET) images from CT scans, patient demographics, and acquisition parameters. The model was trained on 2,538 pseudo-normal PET/CT studies, including stable lymphoma patients and manually disease-masked clinical scans. Model performance was evaluated on 177 test studies achieving 89.3% explained variance and 18.0% mean absolute relative error. We introduced a novel “twin correction” method which reduced SUVmean variance by up to 90% in various organs and successfully reduced confounding normally occurring effects of patient sex, age, fat mass, and uptake time. Finally, anomaly detection and unsupervised tumor segmentation was achieved by comparing actual PET scans with their normal twins. The ntPET-based method achieved a dice score of 49.3% on the AutoPET dataset without requiring tumor annotations for training. In conclusion, the proposed ntPET methodology employs personalized normal references to achieve disease-agnostic patient-specific analysis of PET images.
@article{normaltwin, title = {Normal twin PET: personalized generative modeling for confounder correction and anomaly detection in whole-body PET/CT}, author = {Hinge, Christian and Rodell, Anders Bertil and Zuehlsdorff, Sven and Spottiswoode, Bruce and Korsholm, Kirsten and Fischer, Barbara Malene and Ladefoged, Claes Nøhr and Andersen, Flemming Littrup}, journal = {Scientific Reports}, year = {2025}, month = nov, volume = {15}, publisher = {Nature}, doi = {10.1038/s41598-025-26827-y}, url = {https://www.nature.com/articles/s41598-025-26827-y}, dimensions = {true}, language = {eng}, format = {article}, }
2024
- Diagnostics
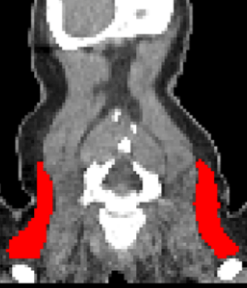 Automated Supraclavicular Brown Adipose Tissue Segmentation in Computed Tomography Using nnU-Net: Integration with TotalSegmentatorKasper Jørgensen, Frederikke Engel Høi-Hansen, Ruth J. F. Loos, and 2 more authorsDiagnostics, Dec 2024
Automated Supraclavicular Brown Adipose Tissue Segmentation in Computed Tomography Using nnU-Net: Integration with TotalSegmentatorKasper Jørgensen, Frederikke Engel Høi-Hansen, Ruth J. F. Loos, and 2 more authorsDiagnostics, Dec 2024Background/Objectives: Brown adipose tissue (BAT) plays a crucial role in energy expenditure and thermoregulation and has thus garnered interest in the context of metabolic diseases. Segmentation in medical imaging is time-consuming and prone to inter- and intra-operator variability. This study aims to develop an automated BAT segmentation method using the nnU-Net deep learning framework, integrated into the TotalSegmentator software, and to evaluate its performance in a large cohort of patients with lymphoma. Methods: A 3D nnU-Net model was trained on the manually annotated BAT regions from 159 lymphoma patients’ CT scans, employing a 5-fold cross-validation approach. An ensemble model was created using these folds to enhance segmentation performance. The model was tested on an independent cohort of 30 patients. The evaluation metrics included the DICE score and Hausdorff Distance (HD). Additionally, the mean standardized uptake value (SUV) in the BAT regions was analyzed in 7107 FDG PET/CT lymphoma studies to identify patterns in the BAT SUVs. Results: The ensemble model achieved a state-of-the-art average DICE score of 0.780 ± 0.077 and an HD of 29.0 ± 14.6 mm in the test set, outperforming the individual fold models. Automated BAT segmentation revealed significant differences in the BAT SUVs between the sexes, with higher values in women. The morning scans showed a higher BAT SUV compared to the afternoon scans, and seasonal variations were observed, with an increased uptake during the winter. The BAT SUVs decreased with age. Conclusions: The proposed automated BAT segmentation tool demonstrates robust performance, reducing the need for manual annotation. The analysis of a large patient cohort confirms the known patterns of BAT SUVs, highlighting the method’s potential for broader clinical and research applications.
@article{batsegmentation, title = {Automated Supraclavicular Brown Adipose Tissue Segmentation in Computed Tomography Using nnU-Net: Integration with TotalSegmentator}, author = {Jørgensen, Kasper and Høi-Hansen, Frederikke Engel and Loos, Ruth J. F. and Hinge, Christian and Andersen, Flemming Littrup}, journal = {Diagnostics}, year = {2024}, month = dec, volume = {14}, issn = {2075-4418}, publisher = {MDPI}, doi = {10.3390/diagnostics14242786}, url = {https://www.mdpi.com/2075-4418/14/24/2786}, dimensions = {true}, language = {eng}, format = {article}, }
2022
- Front. Neurosci.
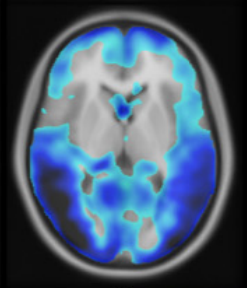 A zero-dose synthetic baseline for the personalized analysis of [18F]FDG-PET: Application in Alzheimer’s diseaseChristian Hinge, Otto Mølby Henriksen, Ulrich Lindberg, and 5 more authorsFrontiers in Neuroscience, Dec 2022
A zero-dose synthetic baseline for the personalized analysis of [18F]FDG-PET: Application in Alzheimer’s diseaseChristian Hinge, Otto Mølby Henriksen, Ulrich Lindberg, and 5 more authorsFrontiers in Neuroscience, Dec 2022Purpose: Brain 2-Deoxy-2-[18F]fluoroglucose ([18F]FDG-PET) is widely used in the diagnostic workup of Alzheimer’s disease (AD). Current tools for uptake analysis rely on non-personalized templates, which poses a challenge as decreased glucose uptake could reflect neuronal dysfunction, or heterogeneous brain morphology associated with normal aging. Overcoming this, we propose a deep learning method for synthesizing a personalized [18F]FDG-PET baseline from the patient’s own MRI, and showcase its applicability in detecting AD pathology. Methods: We included [18F]FDG-PET/MRI data from 123 patients of a local cohort and 600 patients from ADNI. A supervised, adversarial model with two connected Generative Adversarial Networks (GANs) was trained on cognitive normal (CN) patients with transfer-learning to generate full synthetic baseline volumes (sbPET) (192 × 192 × 192) which reflect healthy uptake conditioned on brain anatomy. Synthetic accuracy was measured by absolute relative %-difference (Abs%), relative %-difference (RD%), and peak signal-to-noise ratio (PSNR). Lastly, we deployed the sbPET images in a fully personalized method for localizing metabolic abnormalities. Results: The model achieved a spatially uniform Abs% of 9.4%, RD% of 0.5%, and a PSNR of 26.3 for CN subjects. The sbPET images conformed to the anatomical information dictated by the MRI and proved robust in presence of atrophy. The personalized abnormality method correctly mapped the pathology of AD subjects while showing little to no anomalies for CN subjects. Conclusion: This work demonstrated the feasibility of synthesizing fully personalized, healthy-appearing [18F]FDG-PET images. Using these, we showcased a promising application in diagnosing AD, and theorized the potential value of sbPET images in other neuroimaging routines.
@article{zerodose, title = {A zero-dose synthetic baseline for the personalized analysis of [18F]FDG-PET: Application in Alzheimer's disease}, author = {Hinge, Christian and Henriksen, Otto Mølby and Lindberg, Ulrich and Hasselbalch, Steen Gregers and Højgaard, Liselotte and Law, Ian and Andersen, Flemming Littrup and Ladefoged, Claes Nøhr}, journal = {Frontiers in Neuroscience}, year = {2022}, month = dec, volume = {16}, issn = {1662453x, 16624548}, publisher = {Frontiers Media S.A.}, doi = {10.3389/fnins.2022.1053783}, url = {https://www.frontiersin.org/articles/10.3389/fnins.2022.1053783/full}, dimensions = {true}, language = {eng}, format = {article}, }